Hilary Parker
Hilary Parker
| My primary research interest is in genomics. I work with Jeff Leek as part of the Genomics Working Group. My current project examines the role of batch effects in high-throughput data, specifically microarray data. This issue is especially important because of frequent confounding in experimental designs. |
|
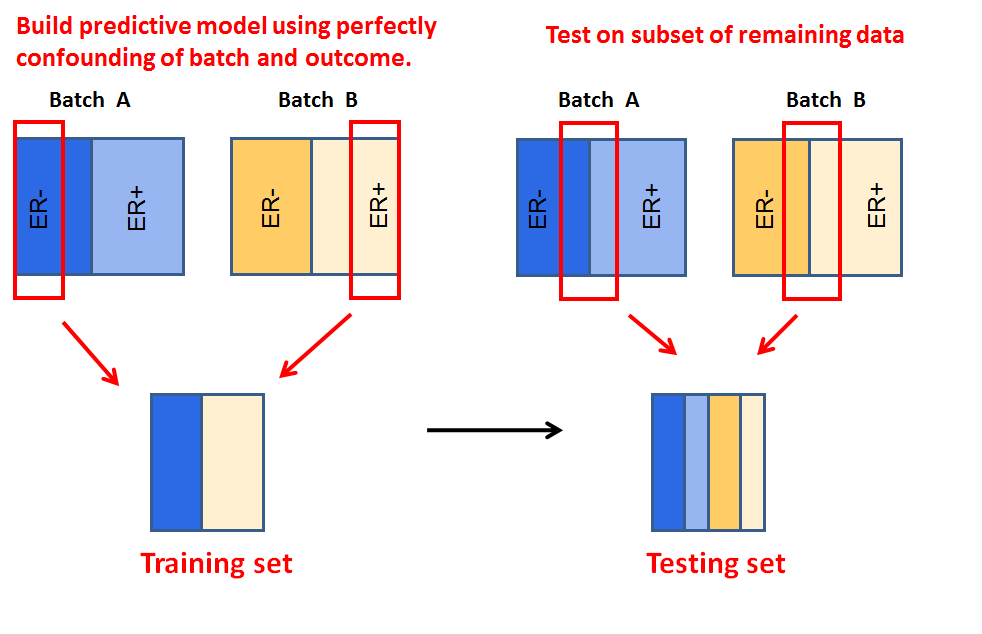
My publication "The Practical Effect of Batch on Genomic Prediction" looks at the role that batch plays when using genomic technologies to perform prediction problems. We found that the correlation between batch and outcome can greatly influence the performance of a predictor. We also found that a subset of effects (in this case microarray probes) drive the negative influence of batch in predictors. |
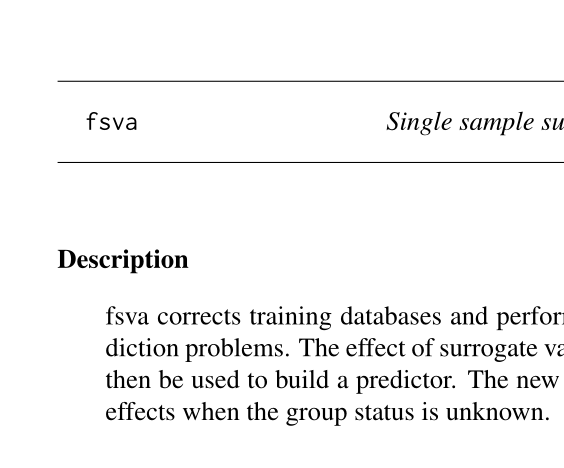
| My current work focuses on updating current batch-effect correction methods to deal with prediction problems for single samples, or confounded samples. This is an important problem in the field of personalized medicine as we begin to see genomic technologies being implemented for diagnostic purposes. The code for our new method, Frozen Surrogate Variable Analysis (fSVA) is currently available in the SVA package on Bioconductor. A publication is in preparation. |
|

|

|

|