
|
Tutorial 6: Determine the Key Motif |
|
Often, from a collection of motifs discovered from ChIP-chip data, we wish to identify which one is the key motif that induces sequence specific protein-DNA finding. For this purpose, we propose to rank motifs according to their relative enrichment levels. In other words, for each motif, we first compute its occurrence rate in ChIP-chip binding regions (or any collection of positive regions where you think the motif should be enriched) as well as its occurrence rate in some negative control regions. We then compare the two occurrence rate to derive a relative fold enrichment. Finally we rank all motifs based on the enrichment. The hypothesis is that the key motif should have the highest enrichment and therefore should rank as the No. 1. Ji et al. (2006) showed that this is generally true, however only when the negative control regions are selected carefully to match physical properties of ChIP-chip binding regions. The paper proposed a simple way to choose matched genomic controls to solve the problem, which has now been incorporated into CisGenome.
To choose matched genomic control regions for a collection of positive regions (e.g., ChIP-chip binding regions), you can first click the menu “Genome > Get Matched Control Region”. |
|
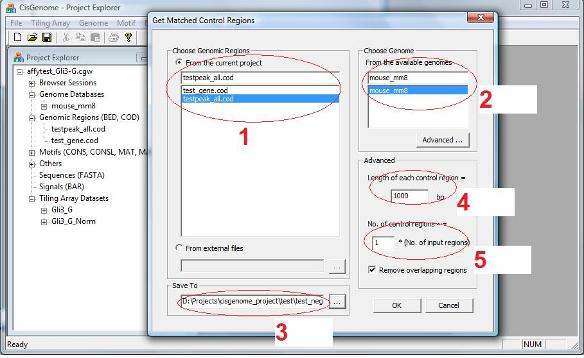
A dialog will jump out. In this dialog, you need to specify: 1. A COD file that contains the coordinates of your positive regions (e.g., ChIP-chip regions, see red circle 1 below); 2. A genome which your coordinates are based on (red circle 2). 3. An output file to save the results (red circle 3). |
|
De novo motif discovery such as Gibbs Motif Sampling usually returns multiple motifs. If you don’t know the motif of your transcription factor, a natural question is “which one of these discovered motif are the one I’m looking for?”. In one of our recent work, we have shown that this can be addressed by comparing motifs’ relative enrichment levels computed using carefully chosen matched genomic control regions. Next we will illustrate how to do this comparison using CisGenome. |
|
You can also specify the length of each negative control region (red circle 4) and how many negative regions to return (red circle 5). When you specify how many regions to return, you can set the number to be one times, two times, … etc. of the number of input positive regions.
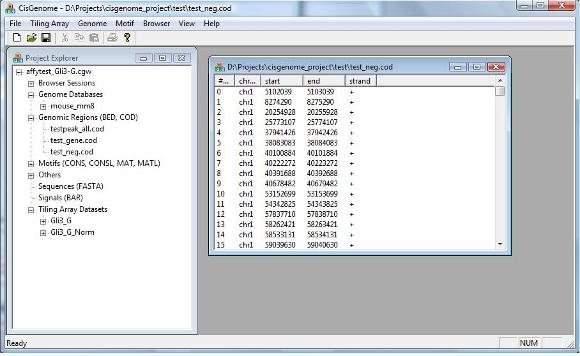
After you finish, click “OK”. Soon you will get a list of negative control regions, listed in a new Window. A COD file that contains these regions will also be added to the Project Explorer window, under the section “Genomic Regions (BED, COD)”. |
1. Choose Matched Genomic Control Regions |
2. Obtain Relative Enrichment Levels for a List of Motifs |
|
Using the same set of positive regions, you can get DNA sequences and perform de novo motif discovery using CisGenome. After you have obtained a list of new motifs and a list of negative control regions, you can now compute relative enrichment levels of these motifs.
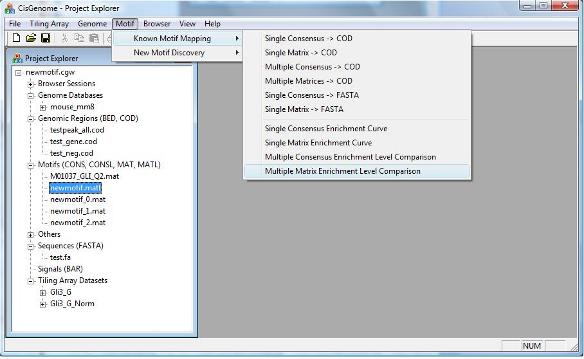
In order to do so, first click menu “Motif > Known Motif Mapping > Multiple Matrix Enrichment Level Comparison”. |
|
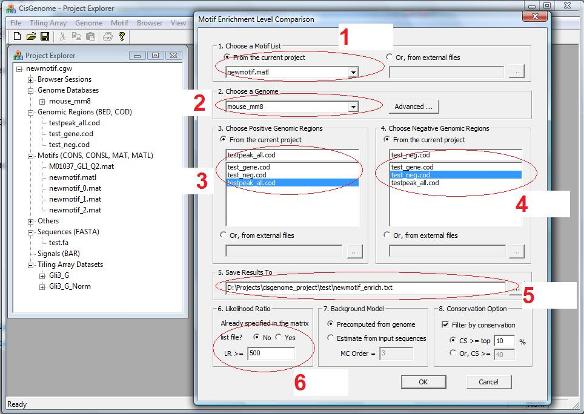
A dialog will show up to let you specify: 1. A motif list (i.e. a *.matl file, see red circle 1 below). Such a file will be generated when you run Gibbs Motif Sampler. 2. A genome which your analysis will be based on (red circle 2). 3. A COD file that contains positive regions (red circle 3). 4. A COD file that contains negative regions (red circle 4). 5. An output file that saves the results (red circle 5). 6. A likelihood ratio cutoff to define TFBS (red circle 6). |
|
You can also choose the type of background models, and you can choose whether the analysis will be repeated for conserved segments within your input regions.
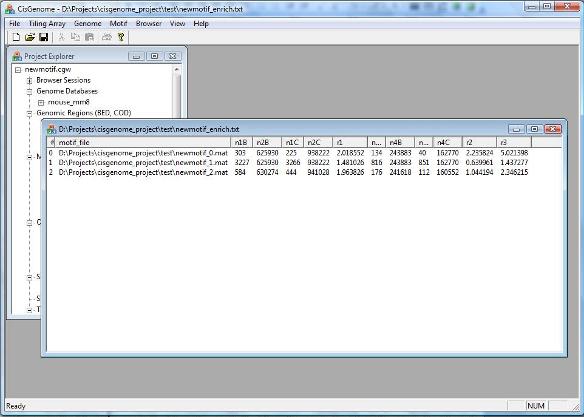
After you finish, click “OK”. After a while, a new window will be opened to show enrichment levels of all motifs. Pick up the top motifs, usually they are the ones that you want to follow up! |
|
Congratulations! At this point, you have already finished the tutorial and becomes an intermediate level user of CisGenome! Now it’s time for you to try it in your own research projects. Good luck! |