
|
Tutorial 6: Novel Motif Discovery |
|
One way to discover new motifs is to search for enriched sequence patterns in transcription factor binding regions identified by ChIP-chip. Such a search can be performed using Gibbs Motif Sampler provided by CisGenome. To run Gibbs motif sampler, first obtain DNA sequences for the target genomic regions and save the sequences into a FASTA file. You can then click the menu “Motif > New Motif Discovery > Gibbs Motif Sampler”. |
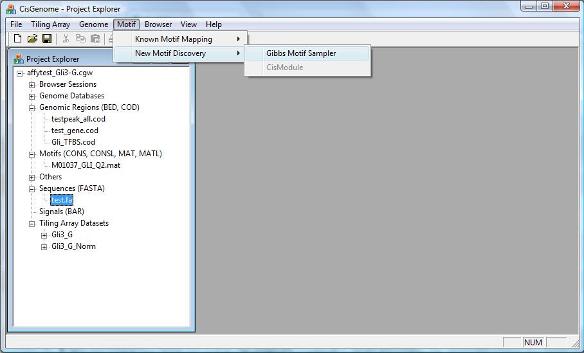
|
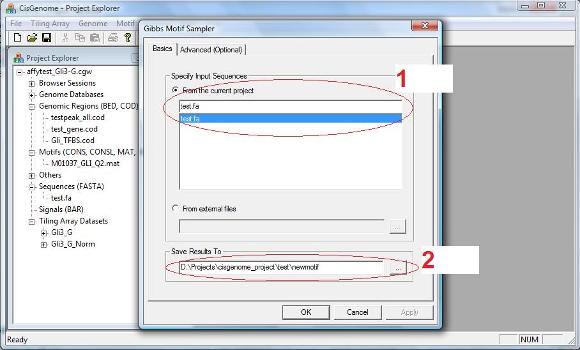
A dialog will jump out. In this dialog, you need to specify: 1. The input sequences (i.e., a FASTA file); 2. An output file (only the file header needs to be specified) to save the results. |
|
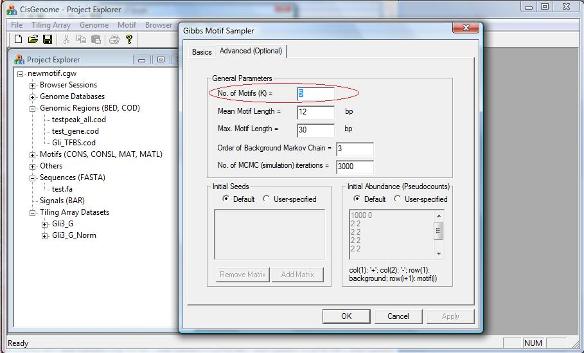
There is an additional page called “Advanced (Optional)” on the dialog. This page allows advanced users to set some optional parameters, e.g., the number of motifs to be searched simultaneously (see red circle below). For the tutorial here, we don’t need to worry about them. |
|
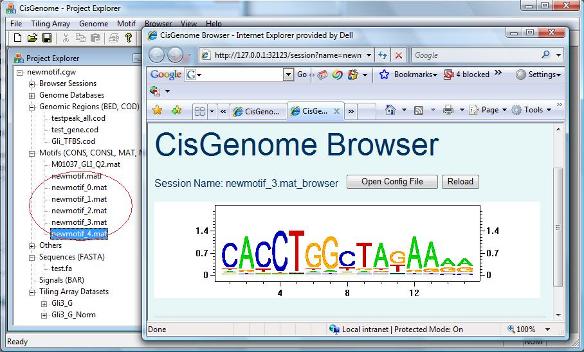
After you click “OK”, the motif sampler will start to scan the sequences. Depending on the length of your input sequence and the number of motifs to be searched, the program may take a few minutes to a few days to run. After it finish, a number of new motif matrices (*.mat) and a list of new motifs (*.matl) will be added to the Project Explorer window, under the section “Motifs (CONS, CONSL, MAT, MATL)” (see red circle below). When you double click the motifs, you will be able to see their sequence logos in CisGenome Browser. |
|
De novo motif discovery such as Gibbs Motif Sampling usually returns multiple motifs. If you don’t know the motif of your transcription factor, a natural question is “which one of these discovered motif are the one I’m looking for?”. In one of our recent work, we have shown that this can be addressed by comparing motifs’ relative enrichment levels computed using carefully chosen matched genomic control regions. Next we will illustrate how to do this comparison using CisGenome. |